Extending Python in C: Modifying NumPy arrays in place
Iterating over masssive amounts of data can be extremely slow in Python. Rewriting performance-critical code in C can improve performance by over 1000x and is easy to do.
Suppose we have an image transformation algorithm that takes an image as input and produces an image as output.
import time
import PIL.Image
import numpy as np
def transform(data, result):
shape = np.shape(data)
data = data / 255
for y in range(1, shape[0]-1):
for x in range(1, shape[1]-1):
v = data[y-1][x-1] * data[y-1][x] * data[y-1][x+1] \
* data[y][x-1] * data[y][x] * data[y][x+1] \
* data[y+1][x-1] * data[y+1][x] * data[y+1][x+1]
d = np.linalg.norm(v)
v = 0 if d == 0.0 else v / d
result[y][x] = np.minimum(v, np.ones(shape[2]))
result *= 255
return result.astype(np.uint8)
def main():
image = PIL.Image.open('test.png')
data = np.array(image)
result = np.empty(np.shape(data))
output = PIL.Image.fromarray(transform(data, result))
output.save('out.png')
if __name__ == '__main__':
main()This code transforms each pixel value into the unit interval by dividing it by 255, multiplies it with the values of all neighbor pixels, normalizes the result and transforms it back into the RGB format by multiplying with 255.
The input image shall be a watermelon pizza.

And the following is the image produced by our algorithm.
A quick benchmark reveals that transforming one image with this algorithm takes over 9 seconds.
> t0 = time.time()
> n = 10
> for _ in range(n):
> main()
> print((time.time() - t0) / n)
9.202898073196412Way too slow. Let’s do some profiling.
> python -m cProfile -s time -o pixel.prof pixel.py
> python -c 'import pstats; p = pstats.Stats("pixel.prof"); p.sort_stats("time").print_stats(4)'
Thu Sep 10 10:41:33 2020 pixel.prof
9214391 function calls (8705887 primitive calls) in 10.604 seconds
Ordered by: internal time
List reduced from 1489 to 4 due to restriction <4>
ncalls tottime percall cumtime percall filename:lineno(function)
1 5.586 5.586 10.329 10.329 pixel.py:5(transform)
505284 1.359 0.000 2.737 0.000 /usr/lib/python3/dist-packages/numpy/linalg/linalg.py:2325(norm)
1515856/1010572 1.177 0.000 3.460 0.000 {built-in method numpy.core._multiarray_umath.implement_array_function}
505286 0.371 0.000 0.371 0.000 {built-in method numpy.empty}
The profiling output reveals that most time is spent indeed in the transform function.
If you prefer a more graphical output, this can be achieved with gprofdot.
> gprof2dot -f pstats pixel.prof | dot -Tpng -o prof.png
This produces the following image.
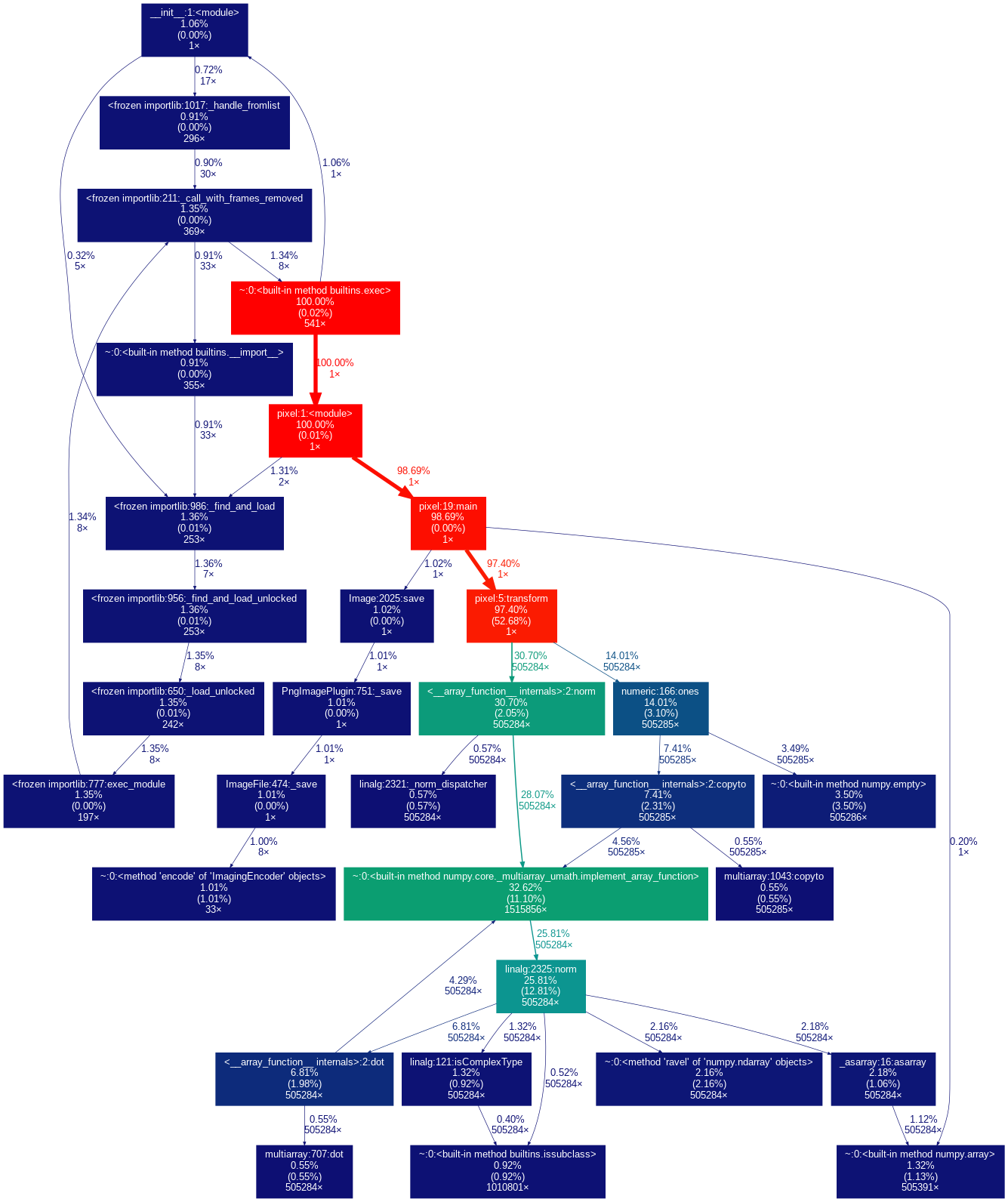
Clearly, the transform function is the problem. It is quickly rewritten in C.
#define NPY_NO_DEPRECATED_API NPY_1_7_API_VERSION
#include <Python.h>
#include <numpy/arrayobject.h>
#include <math.h>
#include <stdbool.h>
inline void set_value(PyArrayObject *data, int y, int x, int z, uint8_t v) {
uint8_t *p = (uint8_t*) PyArray_GETPTR3(data, y, x, z);
*p = v;
}
inline uint8_t get_value(PyArrayObject *data, int y, int x, int z) {
return *((uint8_t*) PyArray_GETPTR3(data, y, x, z));
}
inline void normalize(double *v) {
double imag = 1.0 / sqrt(v[0] * v[0] + v[1] * v[1] + v[2] * v[2]);
v[0] *= imag;
v[1] *= imag;
v[2] *= imag;
}
static PyObject* transform(PyObject* self, PyObject *args) {
PyArrayObject *arrays[2];
if (!PyArg_ParseTuple(args, "O!O!", &PyArray_Type, &arrays[0], &PyArray_Type, &arrays[1])) {
return NULL;
}
int neighbors[][2] = {{-1, -1}, {-1, 0}, {-1, 1},
{0, -1}, {0, 0}, {0, 1},
{1, -1}, {1, 0}, {1, 1}};
long int *shape = PyArray_SHAPE(arrays[0]);
int width=shape[1], height=shape[0];
#pragma omp parallel for
for (int y=1; y<height-1; y++) {
for (int x=1; x<width-1; x++) {
double v[] = {1.0, 1.0, 1.0};
for (int z=0; z<3; z++) {
for(int i=0; i<9; i++) {
v[z] *= get_value(arrays[0], y + neighbors[i][0], x + neighbors[i][1], z) / 255.0;
}
}
normalize(v);
for (int z=0; z<3; z++) {
if (v[z] > 1.0)
v[z] = 1.0;
set_value(arrays[1], y, x, z, v[z] * 255.0);
};
set_value(arrays[1], y, x, 3, 255);
}
}
Py_RETURN_NONE;
}
/* define functions in module */
static PyMethodDef PixelMethods[] =
{
{"transform", transform, METH_VARARGS,
"transform image and write to result array"},
{NULL, NULL, 0, NULL}
};
/* module initialization */
static struct PyModuleDef cModPyDem = {
PyModuleDef_HEAD_INIT,
"_pixel", "transform image",
-1,
PixelMethods
};
PyMODINIT_FUNC PyInit__pixel(void) {
PyObject *module;
module = PyModule_Create(&cModPyDem);
if(module==NULL) return NULL;
/* IMPORTANT: this must be called */
import_array();
if (PyErr_Occurred()) return NULL;
return module;
}To build this C Extension create a setup.py.
from setuptools import setup, Extension
import numpy
setup(
name='pixel',
version='0.1.0',
author='Kris',
author_email='31852063+krisfris@users.noreply.github.com',
description='',
long_description='',
ext_modules=[Extension('_pixel', sources=['pixel.c'],
include_dirs=[numpy.get_include()],
extra_compile_args=['-fopenmp', '-Ofast'],
extra_link_args=['-lgomp'])],
zip_safe=False,
)Then build and install your package.
> python setup.py build
> python setup.py install --user
Or…
> python -m pip install .
If you use poetry, poetry install will automatically build your package and add it to the virtualenv.
Finally, change the main function to use the C Extension.
import _pixel
def main():
image = PIL.Image.open('test.png')
data = np.array(image)
result = np.empty(np.shape(data), dtype=np.uint8)
_pixel.transform(data, result)
output = PIL.Image.fromarray(result)
output.save('out.png')Do another benchmark.
> t0 = time.time()
> n = 10
> for _ in range(n):
> main()
> print((time.time() - t0) / n)
0.143923926353454580.14 seconds. That’s about 64x faster than the Python loop.